|
|
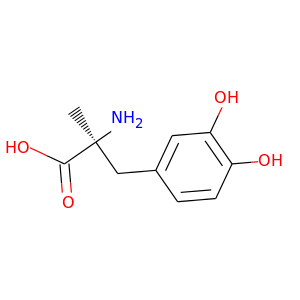
Methyldopa |
211.217 |
 |
|
|
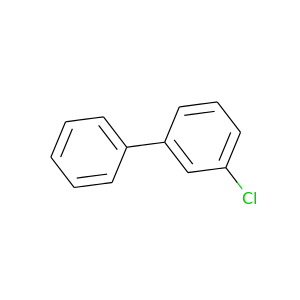
3-Chlorobiphenyl |
188.654 |
 |
|
|
2,2'-Dichlorobiphenyl |
223.096 |
 |
|
|
3,3',4,4'-Tetrachlorobiphenyl |
291.98 |
 |
|
|
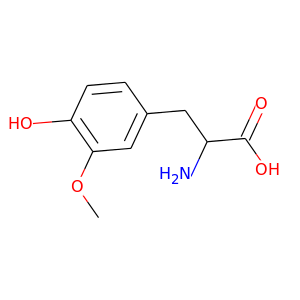
3-Methoxytyrosine |
211.217 |
 |
|
|
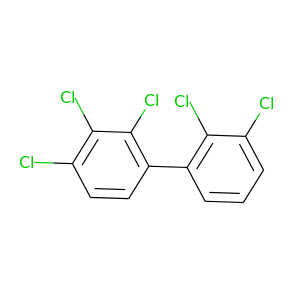
Aroclor 1254 |
326.422 |
 |
|
|
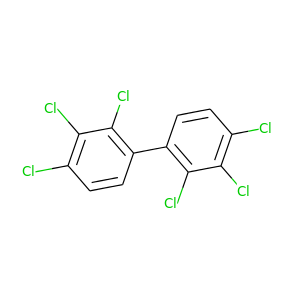
Aroclor 1260 |
360.864 |
 |
|
|
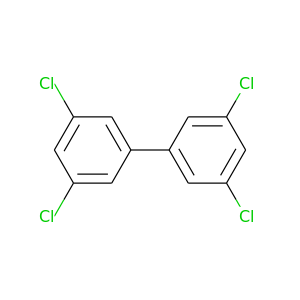
Aroclor 1248 |
291.98 |
 |
|
|
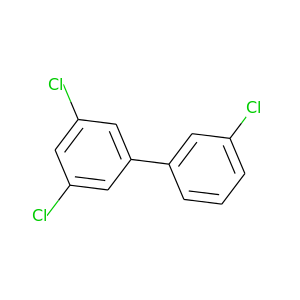
Aroclor 1016 |
257.538 |
 |
|
|
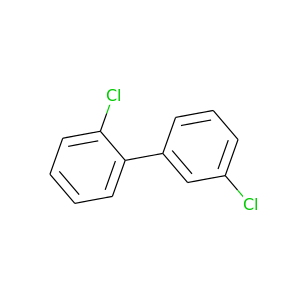
2,3'-Dichlorobiphenyl |
223.096 |
 |
|
|
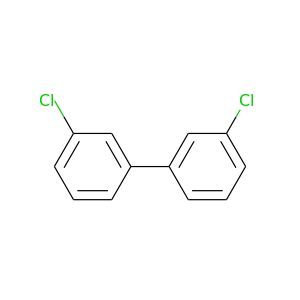
3,3'-Dichlorobiphenyl |
223.096 |
 |
|
|
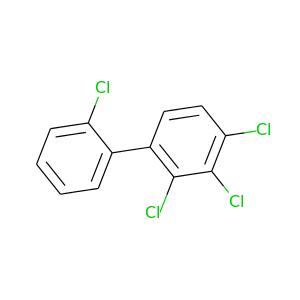
2,2',3,4-Tetrachlorobiphenyl |
291.98 |
 |
|
|
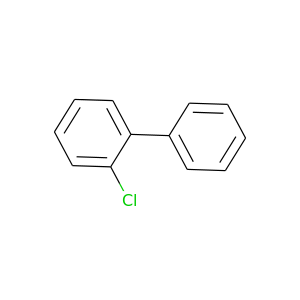
2-Chlorobiphenyl |
188.654 |
 |
|
|
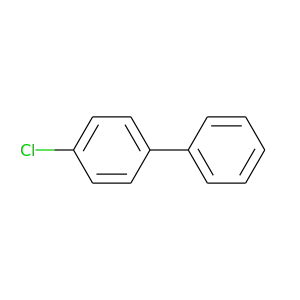
4-Chlorobiphenyl |
188.654 |
 |
|
|
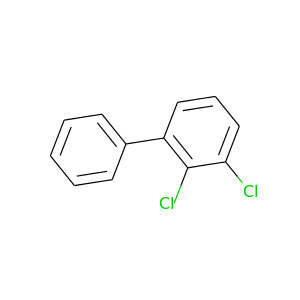
2,3-Dichlorobiphenyl |
223.096 |
 |
|
|
2,4-Dichlorobiphenyl |
223.096 |
 |
|
|
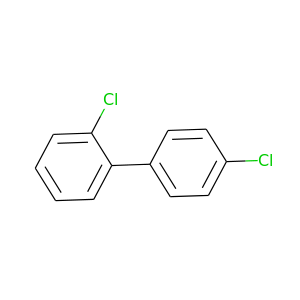
2,4'-Dichlorobiphenyl |
223.096 |
 |
|
|
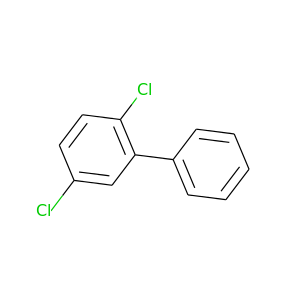
2,5-Dichlorobiphenyl |
223.096 |
 |
|
|
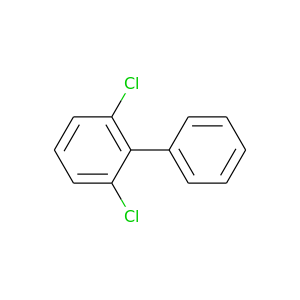
2,6-Dichlorobiphenyl |
223.096 |
 |
|
|
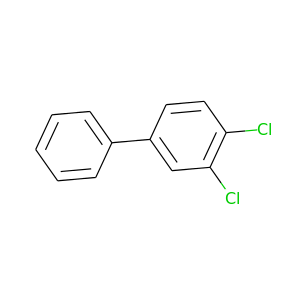
3,4-Dichlorobiphenyl |
223.096 |
 |
|
|
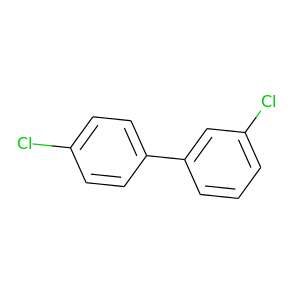
3,4'-Dichlorobiphenyl |
223.096 |
 |
|
|
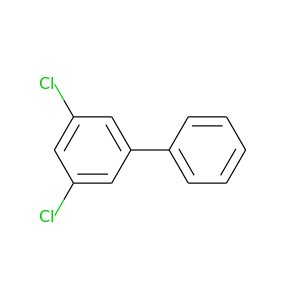
3,5-Dichlorobiphenyl |
223.096 |
 |
|
|
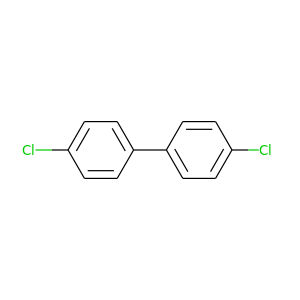
4,4'-Dichlorobiphenyl |
223.096 |
 |
|
|
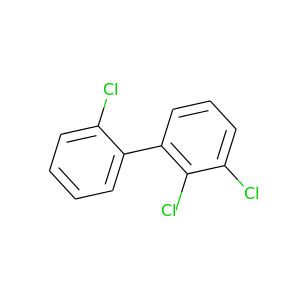
2,2',3-Trichlorobiphenyl |
257.538 |
 |
|
|
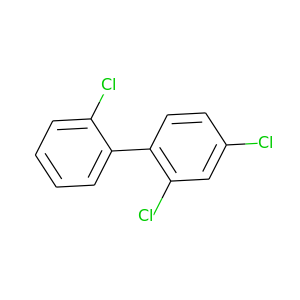
2,2',4-Trichlorobiphenyl |
257.538 |
 |
|
|
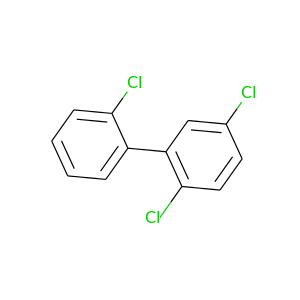
2,2',5-Trichlorobiphenyl |
257.538 |
 |
|
|
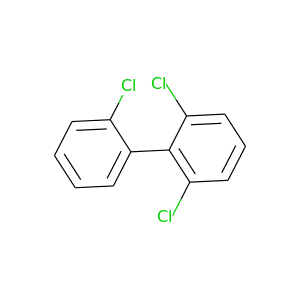
2,2',6-Trichlorobiphenyl |
257.538 |
 |
|
|
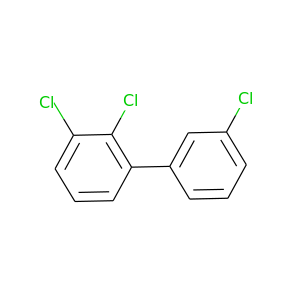
2,3,3'-Trichlorobiphenyl |
257.538 |
 |
|
|
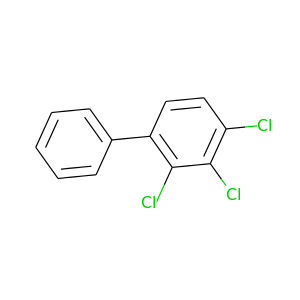
2,3,4-Trichlorobiphenyl |
257.538 |
 |
|
|
3,4,5-Tricholobiphenyl |
257.538 |
 |
|
|
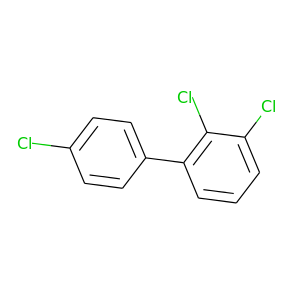
2,3,4'-Trichlorobiphenyl |
257.538 |
 |
|
|
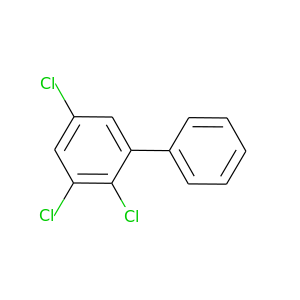
2,3,5-Trichlorobiphenyl |
257.538 |
 |
|
|
2,3,6-Trichlorobiphenyl |
257.538 |
 |
|
|
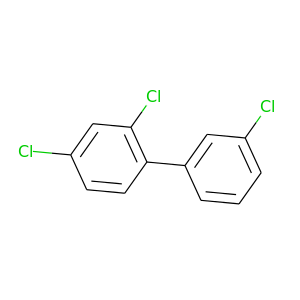
2,3',4-Trichlorobiphenyl |
257.538 |
 |
|
|
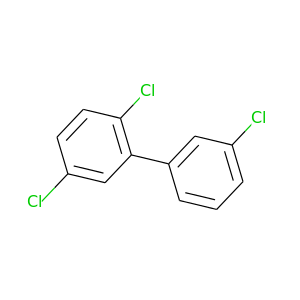
2,3',5-Trichlorobiphenyl |
257.538 |
 |
|
|
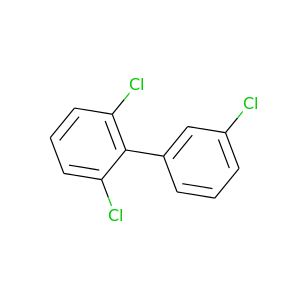
2,3',6-Trichlorobiphenyl |
257.538 |
 |
|
|
2,4,4'-Trichlorobiphenyl |
257.538 |
 |
|
|
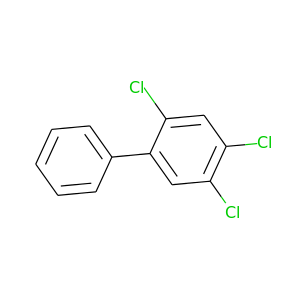
2,4,5-Trichlorobiphenyl |
257.538 |
 |
|
|
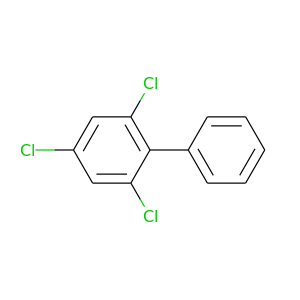
2,4,6-Trichlorobiphenyl |
257.538 |
 |
|
|
2,4',5-Trichlorobiphenyl |
257.538 |
 |
|
|
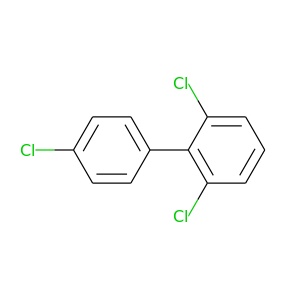
2,4',6-Trichlorobiphenyl |
257.538 |
 |
|
|
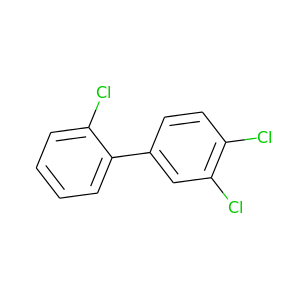
2,3',4'-Trichlorobiphenyl |
257.538 |
 |
|
|
2,3',5'-Trichlorobiphenyl |
257.538 |
 |
|
|
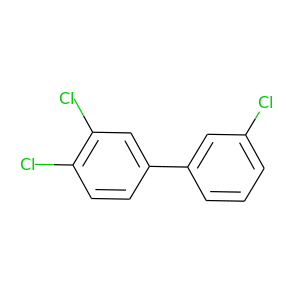
3,3',4-Trichlorobiphenyl |
257.538 |
 |
|
|
3,4,4'-Trichlorobiphenyl |
257.538 |
 |
|
|
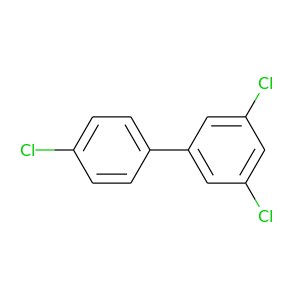
3,4',5-Trichlorobiphenyl |
257.538 |
 |
|
|
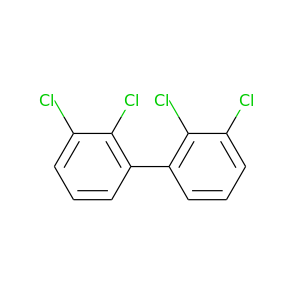
2,2',3,3'-Tetrachlorobiphenyl |
291.98 |
 |
|
|
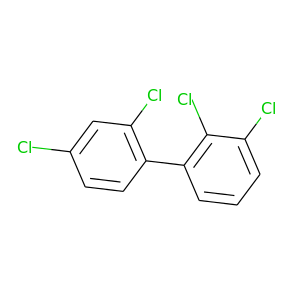
2,2',3,4'-Tetrachlorobiphenyl |
291.98 |
 |
|
|
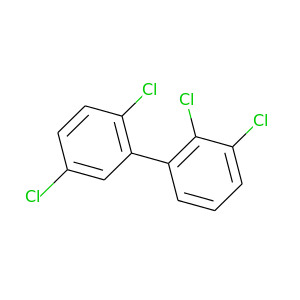
2,2',3,5-Tetrachlorobiphenyl |
291.98 |
 |
|
|
2,2',3,6-Tetrachlorobiphenyl |
291.98 |
 |
|
|
2,2',4,5-Tetrachlorobiphenyl |
291.98 |
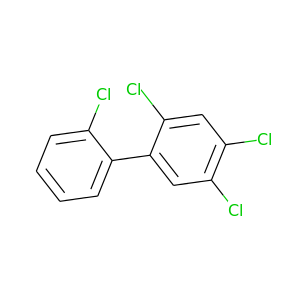 |
|
|
2,2',4,5'-Tetrachlorobiphenyl |
291.98 |
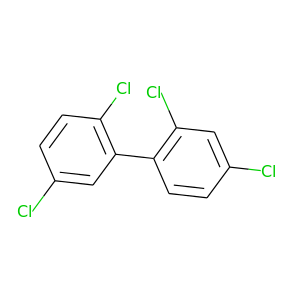 |
|
|
2,2',4,6-Tetrachlorobiphenyl |
291.98 |
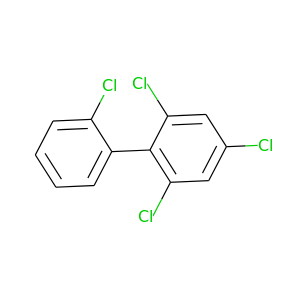 |
|
|
2,2',4,6'-Tetrachlorobiphenyl |
291.98 |
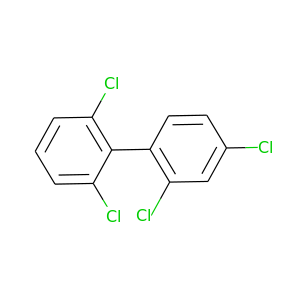 |
|
|
2,2',5,5'-Tetrachlorobiphenyl |
291.98 |
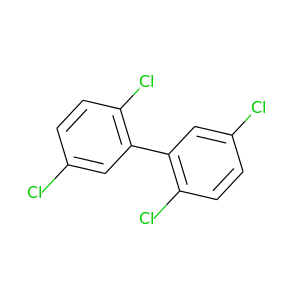 |
|
|
2,2',5,6'-Tetrachlorobiphenyl |
291.98 |
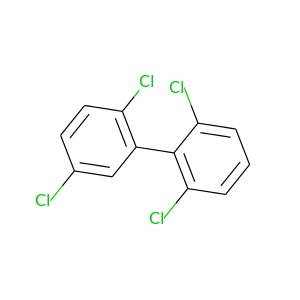 |
|
|
2,2',6,6'-Tetrachlorobiphenyl |
291.98 |
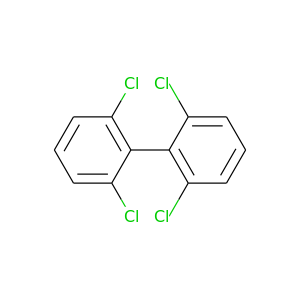 |
|
|
2,3,3',4-Tetrachlorobiphenyl |
291.98 |
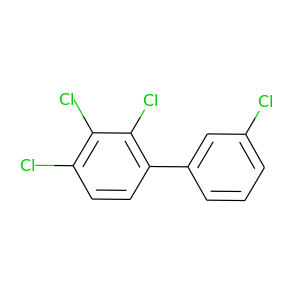 |
|
|
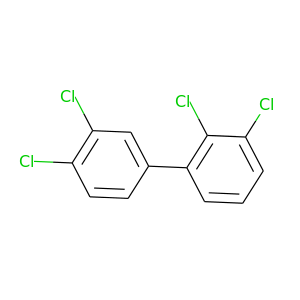
2,3,3',4'-Tetrachlorobiphenyl |
291.98 |
 |
|
|
2,3,3',5-Tetrachlorobiphenyl |
291.98 |
 |
|
|
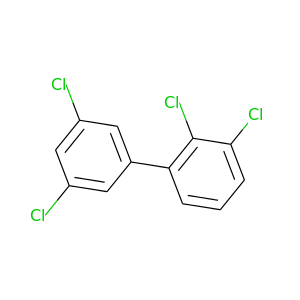
2,3,3',5'-Tetrachlorobiphenyl |
291.98 |
 |
|
|
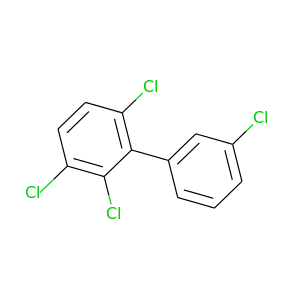
2,3,3',6-Tetrachlorobiphenyl |
291.98 |
 |
|
|
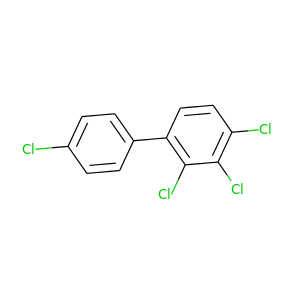
2,3,4,4'-Tetrachlorobiphenyl |
291.98 |
 |
|
|
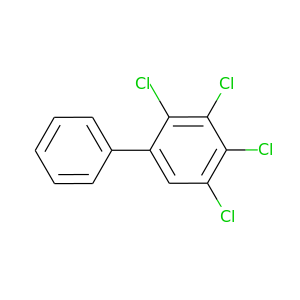
2,3,4,5-Tetrachlorobiphenyl |
291.98 |
 |
|
|
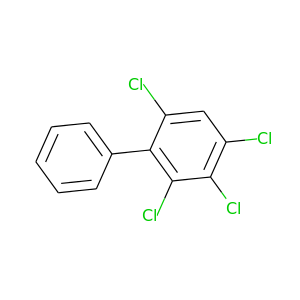
2,3,4,6-Tetrachlorobiphenyl |
291.98 |
 |
|
|
2,3,4',5-Tetrachlorobiphenyl |
291.98 |
 |
|
|
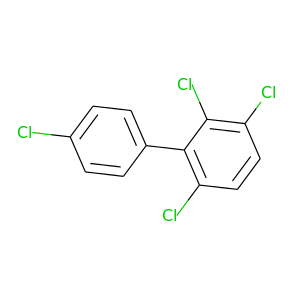
2,3,4',6-Tetrachlorobiphenyl |
291.98 |
 |
|
|
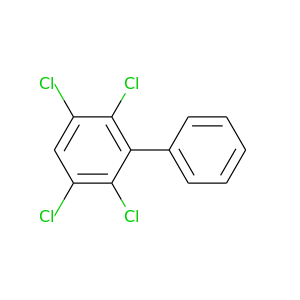
2,3,5,6-Tetrachlorobiphenyl |
291.98 |
 |
|
|
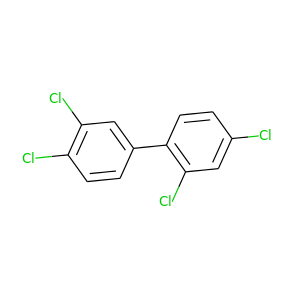
2,3',4,4'-Tetrachlorobiphenyl |
291.98 |
 |
|
|
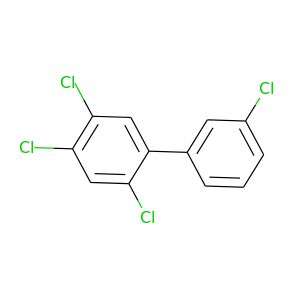
2,3',4,5-Tetrachlorobiphenyl |
291.98 |
 |
|
|
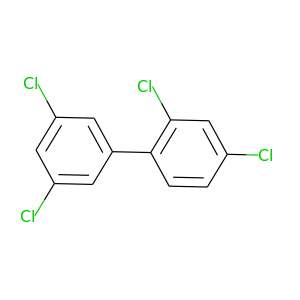
2,3',4,5'-Tetrachlorobiphenyl |
291.98 |
 |
|
|
2,3',4,6-Tetrachlorobiphenyl |
291.98 |
 |
|
|
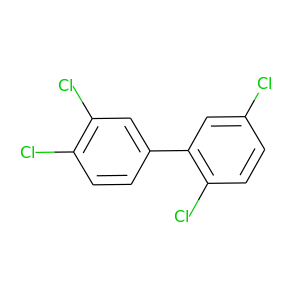
2,3',4',5-Tetrachlorobiphenyl |
291.98 |
 |
|
|
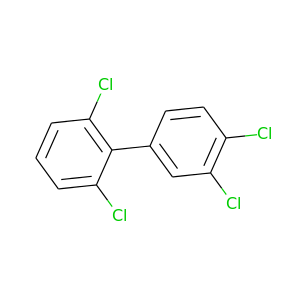
2,3',4',6-Tetrachlorobiphenyl |
291.98 |
 |
|
|
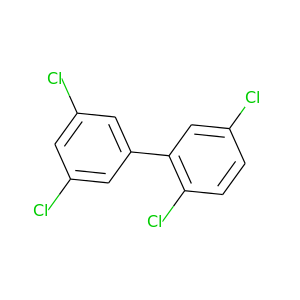
2,3',5,5'-Tetrachlorobiphenyl |
291.98 |
 |
|
|
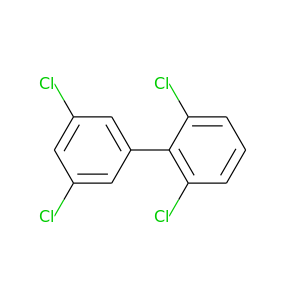
2,3',5',6-Tetrachlorobiphenyl |
291.98 |
 |
|
|
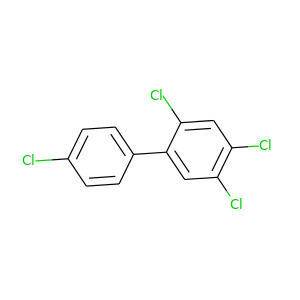
2,4,4',5-Tetrachlorobiphenyl |
291.98 |
 |
|
|
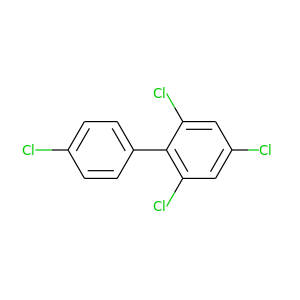
2,4,4',6-Tetrachlorobiphenyl |
291.98 |
 |
|
|
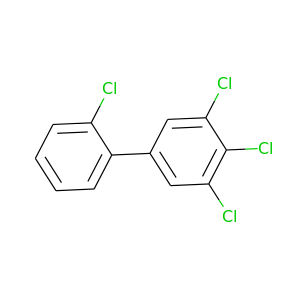
2,3',4',5'-Tetrachlorobiphenyl |
291.98 |
 |
|
|
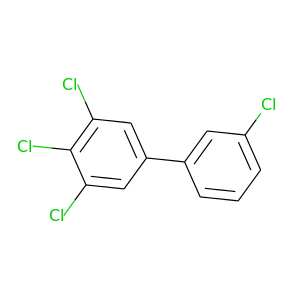
3,3',4,5-Tetrachlorobiphenyl |
291.98 |
 |
|
|
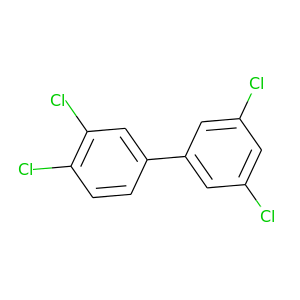
3,3',4,5'-Tetrachlorobiphenyl |
291.98 |
 |
|
|
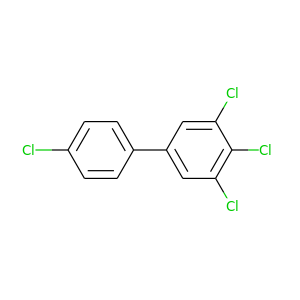
3,4,4',5-Tetrachlorobiphenyl |
291.98 |
 |
|
|
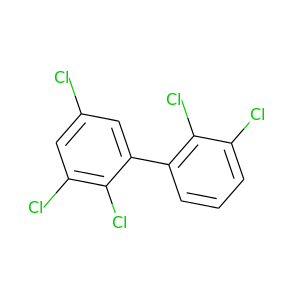
2,2',3,3',5-Pentachlorobiphenyl |
326.422 |
 |
|
|
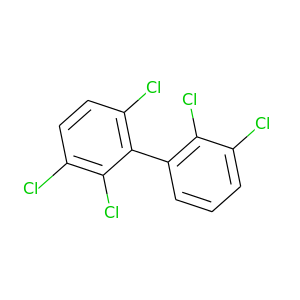
2,2',3,3',6-Pentachlorobiphenyl |
326.422 |
 |
|
|
2,2',3,4,4'-Pentachlorobiphenyl |
326.422 |
 |
|
|
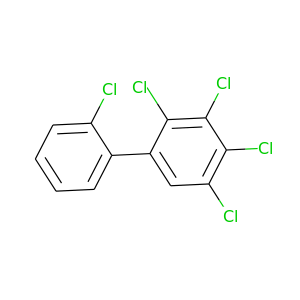
2,2',3,4,5-Pentachlorobiphenyl |
326.422 |
 |
|
|
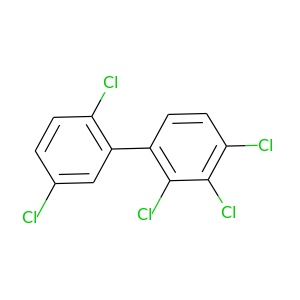
2,2',3,4,5'-Pentachlorobiphenyl |
326.422 |
 |
|
|
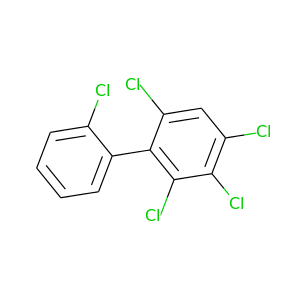
2,2',3,4,6-Pentachlorobiphenyl |
326.422 |
 |
|
|
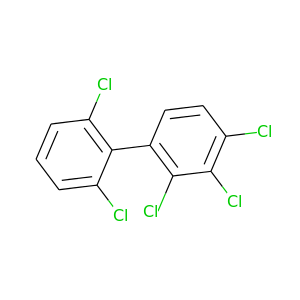
2,2',3,4,6'-Pentachlorobiphenyl |
326.422 |
 |
|
|
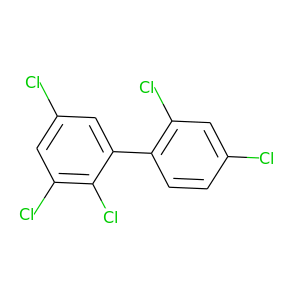
2,2',3,4',5-Pentachlorobiphenyl |
326.422 |
 |
|
|
2,2',3,4',6-Pentachlorobiphenyl |
326.422 |
 |
|
|
2,2',3,5,5'-Pentachlorobiphenyl |
326.422 |
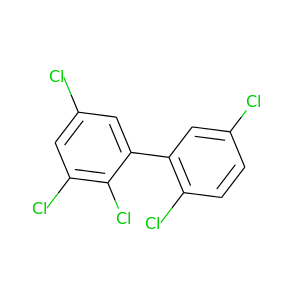 |
|
|
2,2',3,5,6-Pentachlorobiphenyl |
326.422 |
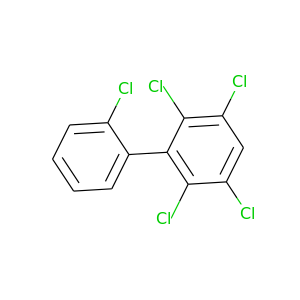 |
|
|
2,2',3,5,6'-Pentachlorobiphenyl |
326.422 |
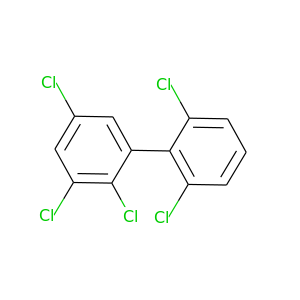 |
|
|
2,2',3,5',6-Pentachlorobiphenyl |
326.422 |
 |
|
|
2,2',3,6,6'-Pentachlorobiphenyl |
326.422 |
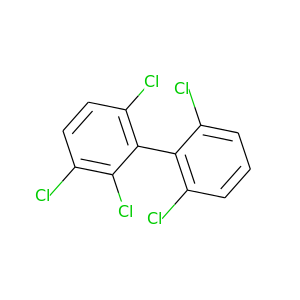 |
|
|
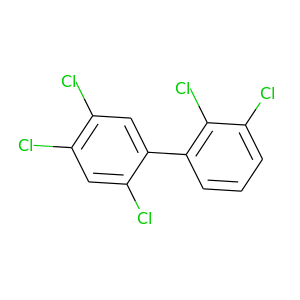
2,2',3,4',5'-Pentachlorobiphenyl |
326.422 |
 |
|
|
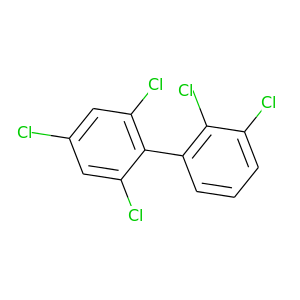
2,2',3,4',6'-Pentachlorobiphenyl |
326.422 |
 |
|
|
2,2',4,4',5-Pentachlorobiphenyl |
326.422 |
 |
|
|
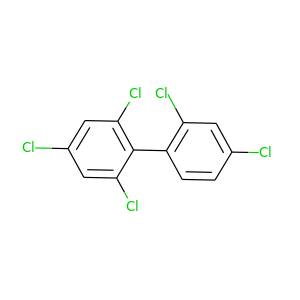
2,2',4,4',6-Pentachlorobiphenyl |
326.422 |
 |
|
|
2,2',4,5,5'-Pentachlorobiphenyl |
326.422 |
 |
|
|
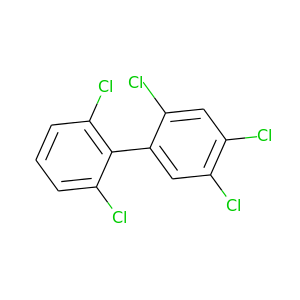
2,2',4,5,6'-Pentachlorobiphenyl |
326.422 |
 |
|
|
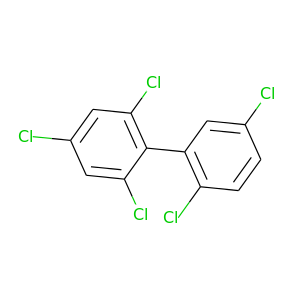
2,2',4,5',6-Pentachlorobiphenyl |
326.422 |
 |
|
|
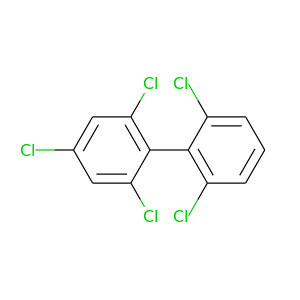
2,2',4,6,6'-Pentachlorobiphenyl |
326.422 |
 |
|
|
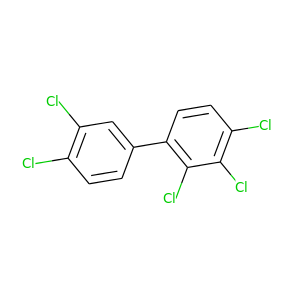
2,3,3',4,4'-Pentachlorobiphenyl |
326.422 |
 |
|
|
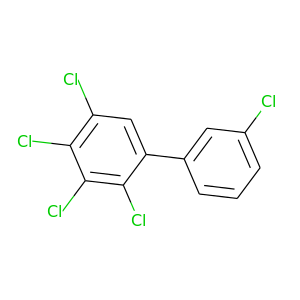
2,3,3',4,5-Pentachlorobiphenyl |
326.422 |
 |
|
|
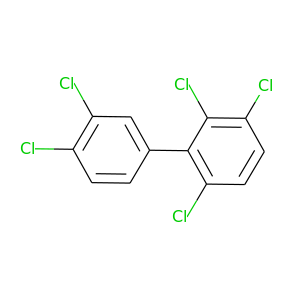
2,3,3',4',5-Pentachlorobiphenyl |
326.422 |
 |
|
|
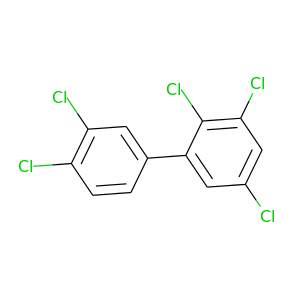
2,3,3',4,5'-Pentachlorobiphenyl |
326.422 |
 |
|
|
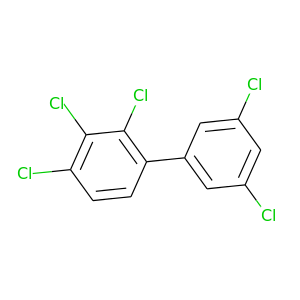
2,3,3',4,6-Pentachlorobiphenyl |
326.422 |
 |
|
|
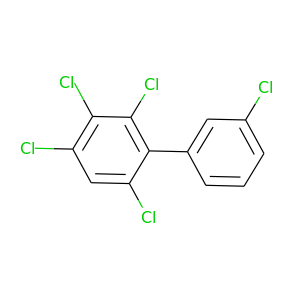
2,3,3',4',6-Pentachlorobiphenyl |
326.422 |
 |
|
|
2,3,3',5,5'-Pentachlorobiphenyl |
326.422 |
 |
|
|
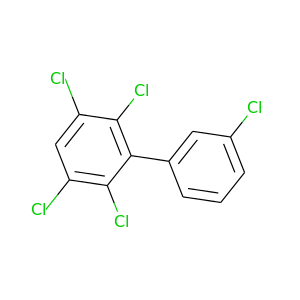
2,3,3',5,6-Pentachlorobiphenyl |
326.422 |
 |
|
|
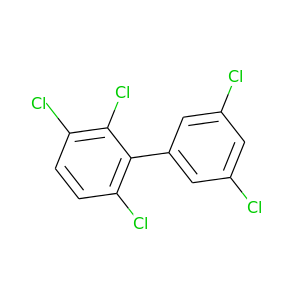
2,3,3',5',6-Pentachlorobiphenyl |
326.422 |
 |
|
|
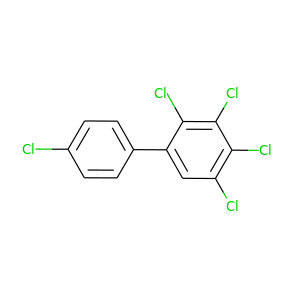
2,3,4,4',5-Pentachlorobiphenyl |
326.422 |
 |
|
|
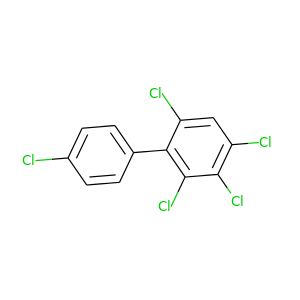
2,3,4,4',6-Pentachlorobiphenyl |
326.422 |
 |
|
|
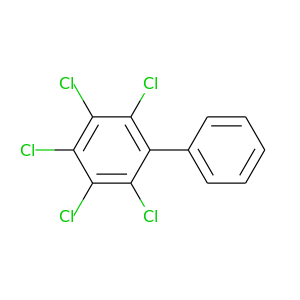
2,3,4,5,6-Pentachlorobiphenyl |
326.422 |
 |
|
|
2,3,4',5,6-Pentachlorobiphenyl |
326.422 |
 |
|
|
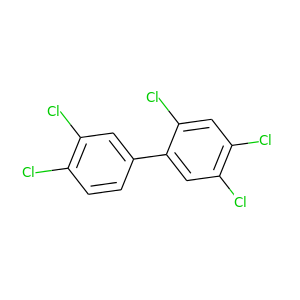
2,3',4,4',5-Pentachlorobiphenyl |
326.422 |
 |
|
|
2,3',4,4',6-Pentachlorobiphenyl |
326.422 |
 |
|
|
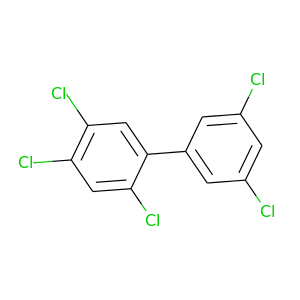
2,3',4,5,5'-Pentachlorobiphenyl |
326.422 |
 |
|
|
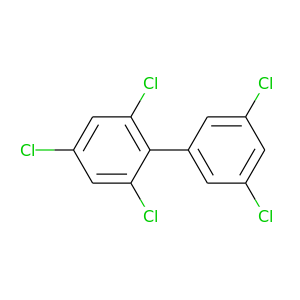
2,3',4,5',6-Pentachlorobiphenyl |
326.422 |
 |
|
|
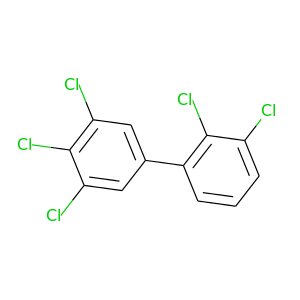
2,3,3',4',5'-Pentachlorobiphenyl |
326.422 |
 |
|
|
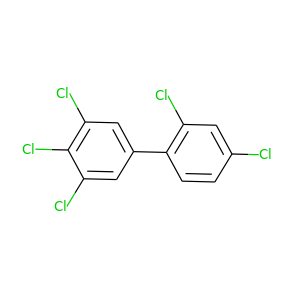
2,3',4,4',5'-Pentachlorobiphenyl |
326.422 |
 |
|
|
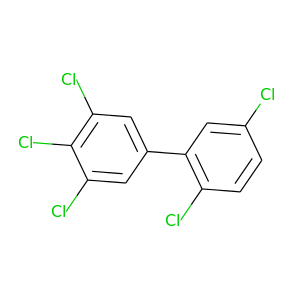
2,3',4',5,5'-Pentachlorobiphenyl |
326.422 |
 |
|
|
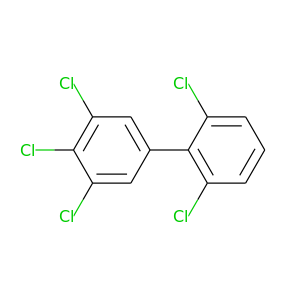
2,3',4',5',6-Pentachlorobiphenyl |
326.422 |
 |
|
|
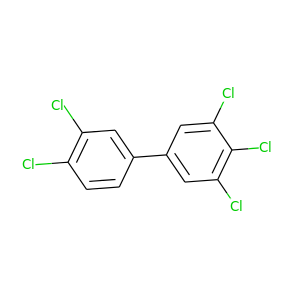
3,3',4,4',5-Pentachlorobiphenyl |
326.422 |
 |
|
|
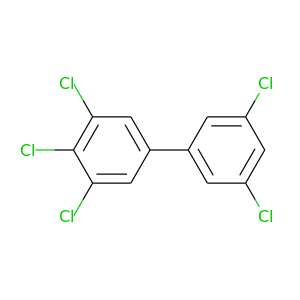
3,3',4,5,5'-Pentachlorobiphenyl |
326.422 |
 |
|
|
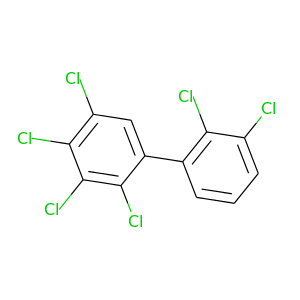
2,2',3,3',4,5-Hexachlorobiphenyl |
360.864 |
 |
|
|
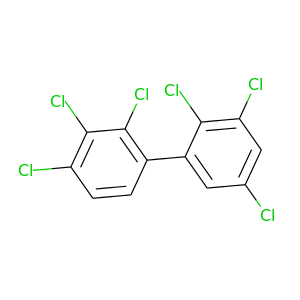
2,2',3,3',4,5'-Hexachlorobiphenyl |
360.864 |
 |
|
|
2,2',3,3',4,6-Hexachlorobiphenyl |
360.864 |
 |
|
|
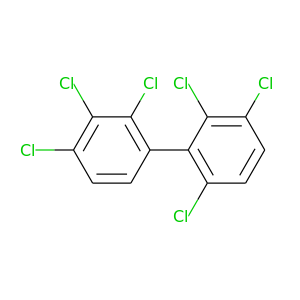
2,2',3,3',4,6'-Hexachlorobiphenyl |
360.864 |
 |
|
|
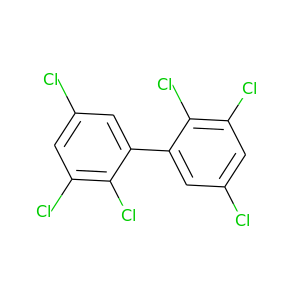
2,2',3,3',5,5'-Hexachlorobiphenyl |
360.864 |
 |
|
|
2,2',3,3',5,6-Hexachlorobiphenyl |
360.864 |
 |
|
|
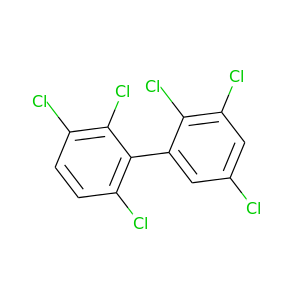
2,2',3,3',5,6'-Hexachlorobiphenyl |
360.864 |
 |
|
|
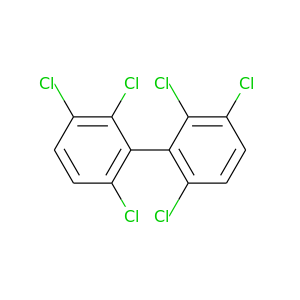
2,2',3,3',6,6'-Hexachlorobiphenyl |
360.864 |
 |
|
|
2,2',3,4,4',5-Hexachlorobiphenyl |
360.864 |
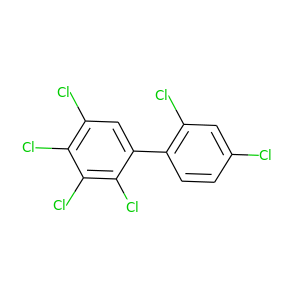 |
|
|
2,2',3,4,4',5'-Hexachlorobiphenyl |
360.864 |
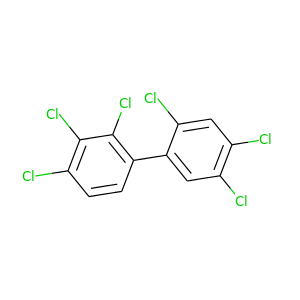 |
|
|
2,2',3,4,4',6-Hexachlorobiphenyl |
360.864 |
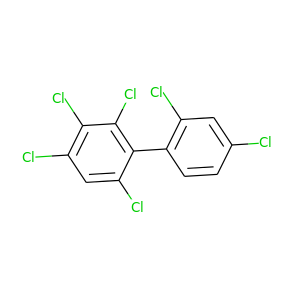 |
|
|
2,2',3,4,4',6'-Hexachlorobiphenyl |
360.864 |
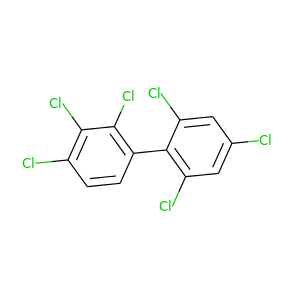 |
|
|
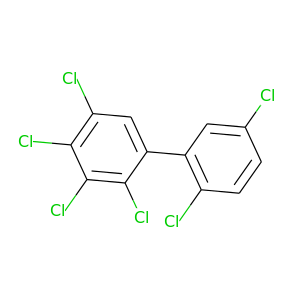
2,2',3,4,5,5'-Hexachlorobiphenyl |
360.864 |
 |
|
|
2,2',3,4,5,6-Hexachlorobiphenyl |
360.864 |
 |
|
|
2,2',3,4,5,6'-Hexachlorobiphenyl |
360.864 |
 |
|
|
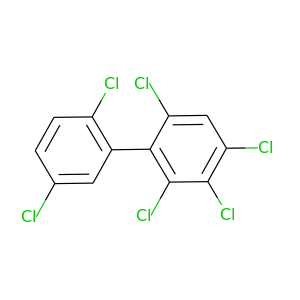
2,2',3,4,5',6-Hexachlorobiphenyl |
360.864 |
 |
|
|
2,2',3,4,6,6'-Hexachlorobiphenyl |
360.864 |
 |
|
|
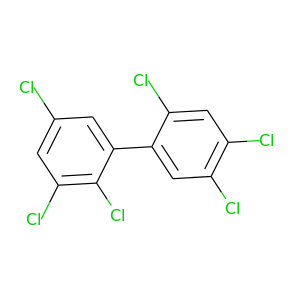
2,2',3,4',5,5'-Hexachlorobiphenyl |
360.864 |
 |
|
|
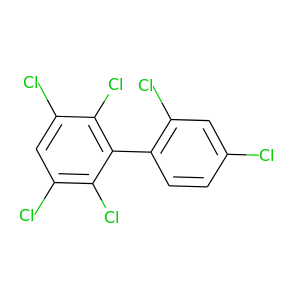
2,2',3,4',5,6-Hexachlorobiphenyl |
360.864 |
 |
|
|
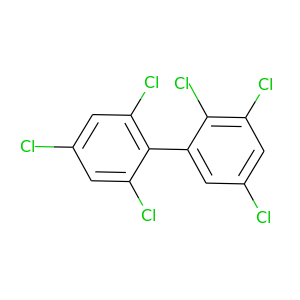
2,2',3,4',5,6'-Hexachlorobiphenyl |
360.864 |
 |
|
|
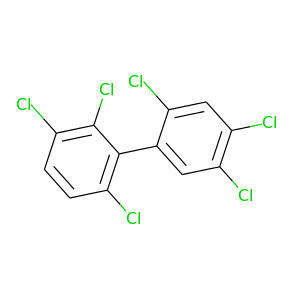
2,2',3,4',5',6-Hexachlorobiphenyl |
360.864 |
 |
|
|
2,2',3,4',6,6'-Hexachlorobiphenyl |
360.864 |
 |
|
|
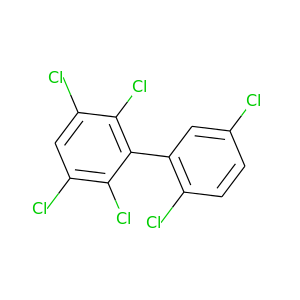
2,2',3,5,5',6-Hexachlorobiphenyl |
360.864 |
 |
|
|
2,2',3,5,6,6'-Hexachlorobiphenyl |
360.864 |
 |
|
|
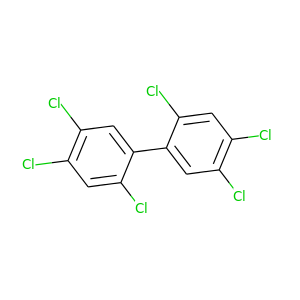
2,2',4,4',5,5'-Hexachlorobiphenyl |
360.864 |
 |
|
|
2,2',4,4',5,6'-Hexachlorobiphenyl |
360.864 |
 |
|
|
2,2',4,4',6,6'-Hexachlorobiphenyl |
360.864 |
 |
|
|
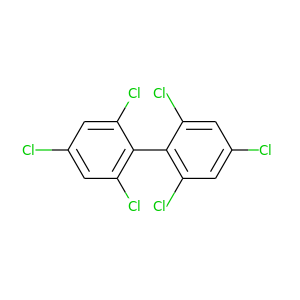
2,3,3',4,4',5-Hexachlorobiphenyl |
360.864 |
 |
|
|
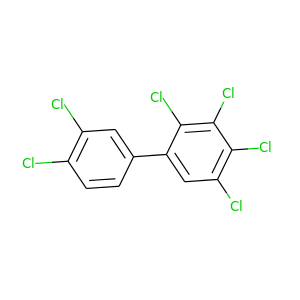
2,3,3',4,4',5'-Hexachlorobiphenyl |
360.864 |
 |
|
|
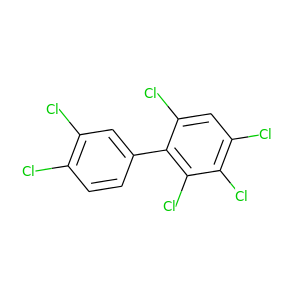
2,3,3',4,4',6-Hexachlorobiphenyl |
360.864 |
 |
|
|
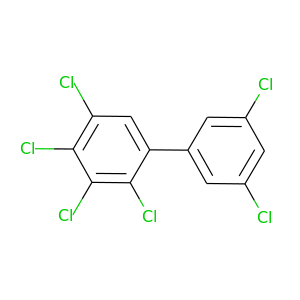
2,3,3',4,5,5'-Hexachlorobiphenyl |
360.864 |
 |
|
|
2,3,3',4,5,6-Hexachlorobiphenyl |
360.864 |
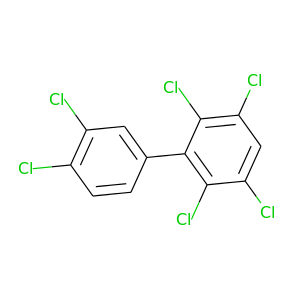 |
|
|
2,3,3',4,5',6-Hexachlorobiphenyl |
360.864 |
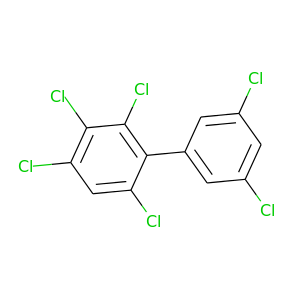 |
|
|
2,3,3',4',5,5'-Hexachlorobiphenyl |
360.864 |
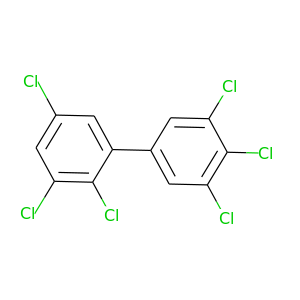 |
|
|
2,3,3',4',5',6-Hexachlorobiphenyl |
360.864 |
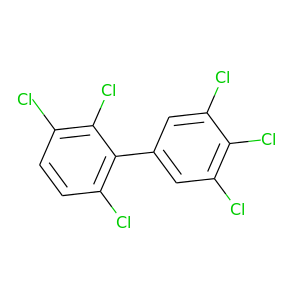 |
|
|
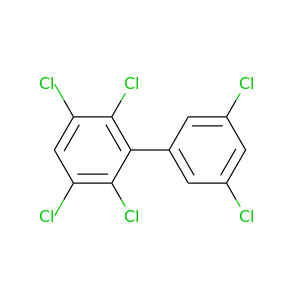
2,3,3',5,5',6-Hexachlorobiphenyl |
360.864 |
 |
|
|
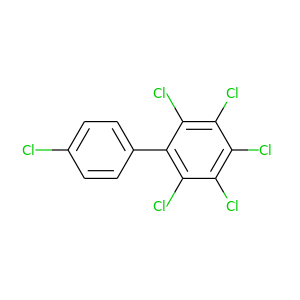
2,3,4,4',5,6-Hexachlorobiphenyl |
360.864 |
 |
|
|
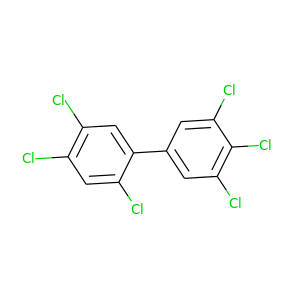
2,3',4,4',5,5'-Hexachlorobiphenyl |
360.864 |
 |
|
|
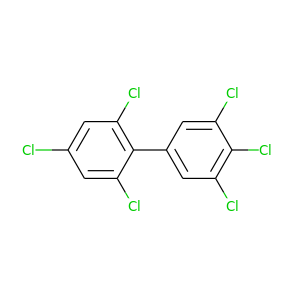
2,3',4,4',5',6-Hexachlorobiphenyl |
360.864 |
 |
|
|
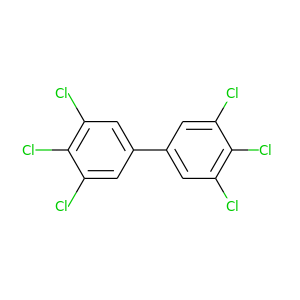
3,3',4,4',5,5'-Hexachlorobiphenyl |
360.864 |
 |
|
|
2,2',3,3',4,4',5-Heptachlorobiphenyl |
395.306 |
 |
|
|
2,2',3,3',4,4',6-Heptachlorobiphenyl |
395.306 |
 |
|
|
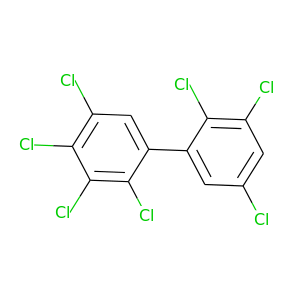
2,2',3,3',4,5,5'-Heptachlorobiphenyl |
395.306 |
 |
|
|
2,2',3,3',4,5,6-Heptachlorobiphenyl |
395.306 |
 |
|
|
2,2',3,3',4,5,6'-Heptachlorobiphenyl |
395.306 |
 |
|
|
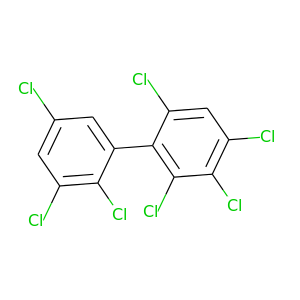
2,2',3,3',4,5',6-Heptachlorobiphenyl |
395.306 |
 |
|
|
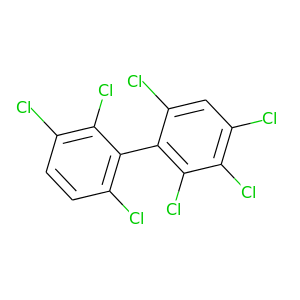
2,2',3,3',4,6,6'-Heptachlorobiphenyl |
395.306 |
 |
|
|
2,2',3,3',4,5',6'-Heptachlorobiphenyl |
395.306 |
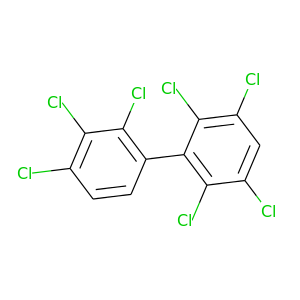 |
|
|
2,2',3,3',5,5',6-Heptachlorobiphenyl |
395.306 |
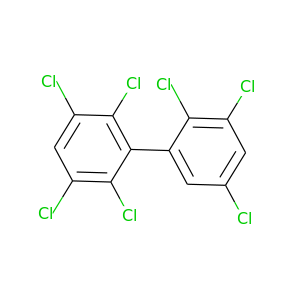 |
|
|
2,2',3,3',5,6,6'-Heptachlorobiphenyl |
395.306 |
 |
|
|
2,2',3,4,4',5,5'-Heptachlorobiphenyl |
395.306 |
 |
|
|
2,2',3,4,4',5,6-Heptachlorobiphenyl |
395.306 |
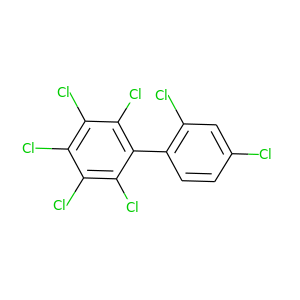 |
|
|
2,2',3,4,4',5,6'-Heptachlorobiphenyl |
395.306 |
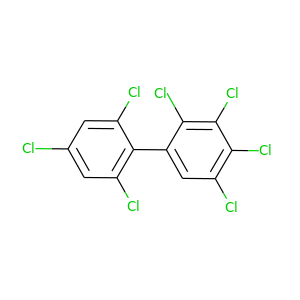 |
|
|
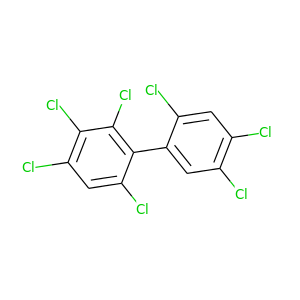
2,2',3,4,4',5',6-Heptachlorobiphenyl |
395.306 |
 |
|
|
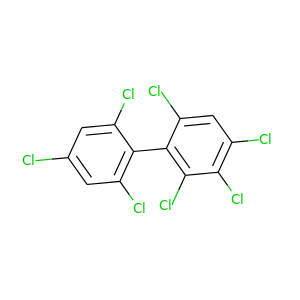
2,2',3,4,4',6,6'-Heptachlorobiphenyl |
395.306 |
 |
|
|
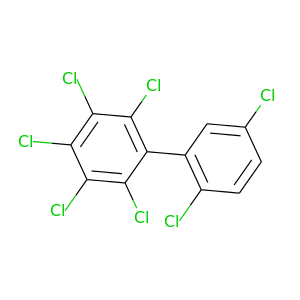
2,2',3,4,5,5',6-Heptachlorobiphenyl |
395.306 |
 |
|
|
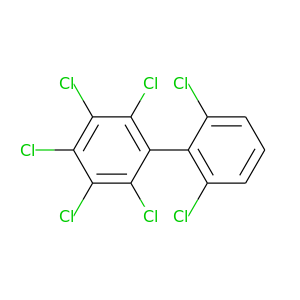
2,2',3,4,5,6,6'-Heptachlorobiphenyl |
395.306 |
 |
|
|
2,2',3,4',5,5',6-Heptachlorobiphenyl |
395.306 |
 |
|
|
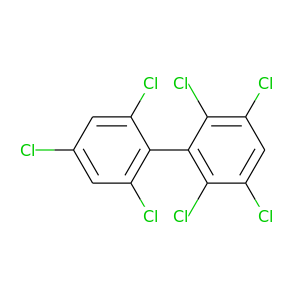
2,2',3,4',5,6,6'-Heptachlorobiphenyl |
395.306 |
 |
|
|
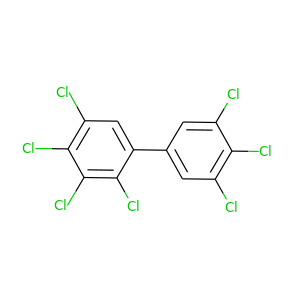
2,3,3',4,4',5,5'-Heptachlorobiphenyl |
395.306 |
 |
|
|
2,3,3',4,4',5,6-Heptachlorobiphenyl |
395.306 |
 |
|
|
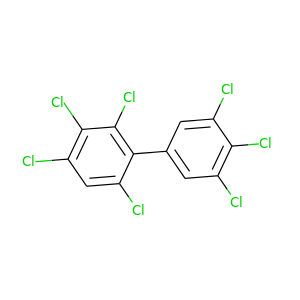
2,3,3',4,4',5',6-Heptachlorobiphenyl |
395.306 |
 |
|
|
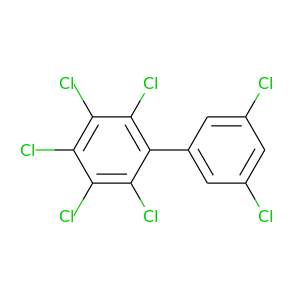
2,3,3',4,5,5',6-Heptachlorobiphenyl |
395.306 |
 |
|
|
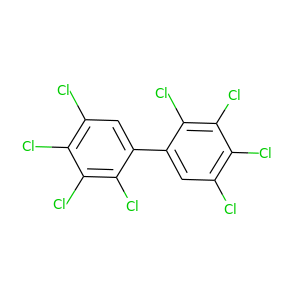
2,2',3,3',4,4',5,5'-Octachlorobiphenyl |
429.748 |
 |
|
|
2,2',3,3',4,4',5,6-Octachlorobiphenyl |
429.748 |
 |
|
|
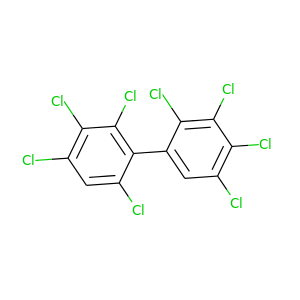
2,2',3,3',4,4',5,6'-Octachlorobiphenyl |
429.748 |
 |
|
|
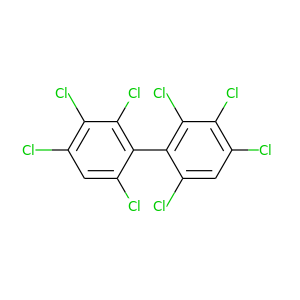
2,2',3,3',4,4',6,6'-Octachlorobiphenyl |
429.748 |
 |
|
|
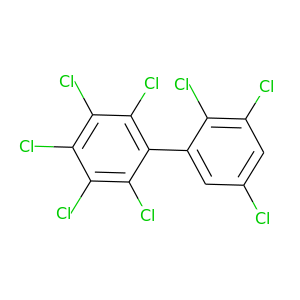
2,2',3,3',4,5,5',6-Octachlorobiphenyl |
429.748 |
 |
|
|
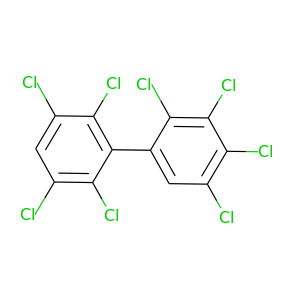
2,2',3,3',4,5,5',6'-Octachlorobiphenyl |
429.748 |
 |
|
|
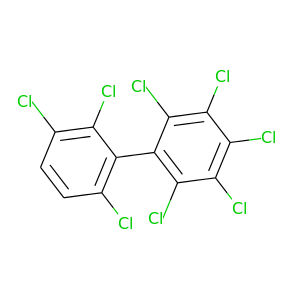
2,2',3,3',4,5,6,6'-Octachlorobiphenyl |
429.748 |
 |
|
|
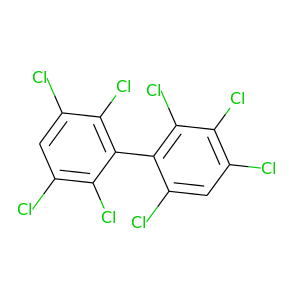
2,2',3,3',4,5',6,6'-Octachlorobiphenyl |
429.748 |
 |
|
|
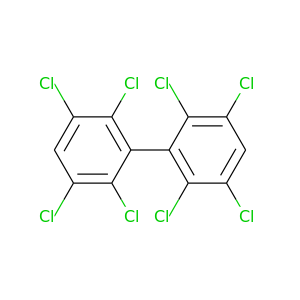
2,2',3,3',5,5',6,6'-Octachlorobiphenyl |
429.748 |
 |
|
|
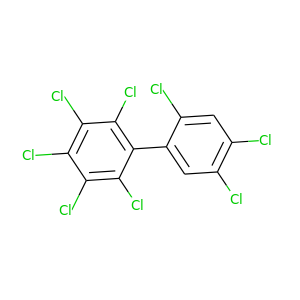
2,2',3,4,4',5,5',6-Octachlorobiphenyl |
429.748 |
 |
|
|
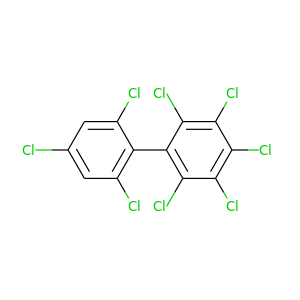
2,2',3,4,4',5,6,6'-Octachlorobiphenyl |
429.748 |
 |
|
|
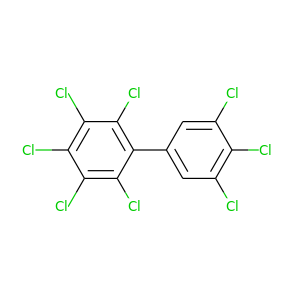
2,3,3',4,4',5,5',6-Octachlorobiphenyl |
429.748 |
 |
|
|
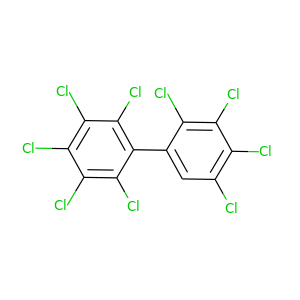
2,2',3,3',4,4',5,5',6-Nonachlorobiphenyl |
464.19 |
 |
|
|
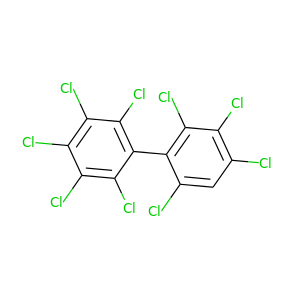
2,2',3,3',4,4',5,6,6'-Nonachlorobiphenyl |
464.19 |
 |
|
|
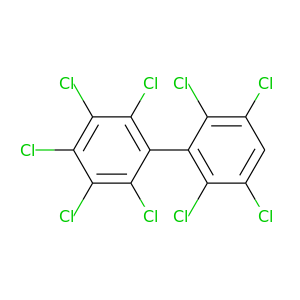
2,2',3,3',4,5,5',6,6'-Nonachlorobiphenyl |
464.19 |
 |
|
|
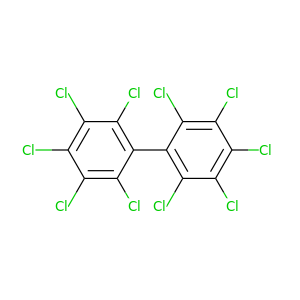
Decachlorobiphenyl |
498.632 |
 |