E.coli Website Introduction:
This 3D environment is designed to present a complete view of metabolite network for biologists, and help them make use of metabolite network of E.coli. It would be full of vitality with constant maintenance and update, to provide great help in drug design and analysis of metabolism.
1. Can't visit or get a disordered page
The page should be loaded as the following picture
If you get the following view, probably because your browser does not support webGL

In order to make your usage more easy and time-saving, we strongly recommend you to use the
Google Chrome web browser (windows platform) or
firefox browser
(Linux platform (Debian/Ubuntu/Fedora/openSUSE)) when visiting
E.coli website.
More Details
Front-end interface is developed based on HTML5 and css3.
3D environment and Real-time Rendering is constructed with WebGL and Javascript.
So your browser should support these new features.
In a word, you should have Javascript(JS) enabled in your browser and make sure that your broswer support HTML, CSS3, WebGL.
Not understand?No problem, just try to use google chrome browser (windows platform) or Firefox (Linux platform) to visit E.coli website. [CLOSE]
In a word, you should have Javascript(JS) enabled in your browser and make sure that your broswer support HTML, CSS3, WebGL.
Not understand?No problem, just try to use google chrome browser (windows platform) or Firefox (Linux platform) to visit E.coli website. [CLOSE]
2. Basic Usages and instructions
2.1. Scroll down to zoom out, scroll up to zoom in.
2.2. Left click, hold, and drag to move the view.
2.3. Mouse over a particular node: the float panels show information that contains pathway, compound name, and pathway link(s).
2.4. Click a node or an edge to get more information.
2.4.1 load structure: load structure picture of selected compound.
2.4.2 related links: related links of this compound(reaction or pathway) to other websites
2.2. Left click, hold, and drag to move the view.
2.3. Mouse over a particular node: the float panels show information that contains pathway, compound name, and pathway link(s).
2.4. Click a node or an edge to get more information.
2.4.1 load structure: load structure picture of selected compound.
2.4.2 related links: related links of this compound(reaction or pathway) to other websites
3. Options Menu
You can get different views by checking the appropriate check box.
3.1. Hide main reactions: Select to hide main reactions.
3.2. Hide main compounds: Select to hide main compounds.
3.3. Show currency compounds: Select to show currency compounds
3.4. Show pathway information: Select to show pathway information when mouse move
3.5. When compounds fade out:
3.5.1 Compounds can be clicked: when you search, some highlight nodes can't be clicked easily, NOT select this to make it easier.
3.1. Hide main reactions: Select to hide main reactions.
3.2. Hide main compounds: Select to hide main compounds.
3.3. Show currency compounds: Select to show currency compounds
3.4. Show pathway information: Select to show pathway information when mouse move
3.5. When compounds fade out:
3.5.1 Compounds can be clicked: when you search, some highlight nodes can't be clicked easily, NOT select this to make it easier.
4. Search Menu
4.1 Search:
Choose category first, then select name or id, at last click search button.
You can also input in text area, then search.
4.2 Search by smiles:
Supporting substructure search.
Input a smile string(for example:c1ccccc1), then search, you can get all compounds include the
sub-structure. Notice the yellow tips which tells you what server is doing.
4.2.1 JSME: a JavaScript Molecular Editor, you can draw a structure and get smiles on line.
4.2.2 Get Smiles: Convert the structure in JSME and fill into text area.
4.2.3 Get Smiles & Search: Get smiles and search by smiles are integrated together.
4.2.1 JSME: a JavaScript Molecular Editor, you can draw a structure and get smiles on line.
4.2.2 Get Smiles: Convert the structure in JSME and fill into text area.
4.2.3 Get Smiles & Search: Get smiles and search by smiles are integrated together.
4.3 Protein Sequence Similarity Search:
You will get several most similar results.
sequence format:
sequence format:
>P0A6B4
MQAATVVINRRALRHNLQRLRELAPASKMVAVVKANAYGHGLLETARTLP
DADAFGVARLEEALRLRAGGITKPVLLLEGFFDARDLPTISAQHFHTAVH
NEEQLAALEEASLDEPVTVWMKLDTGMHRLGVRPEQAEAFYHRLTQCKNV
RQPVNIVSHFARADEPKCGATEKQLAIFNTFCEGKPGQRSIAASGGILLW
PQSHFDWVRPGIILYGVSPLEDRSTGADFGCQPVMSLTSSLIAVREHKAG
Z-score: score via FASTA algorithm.MQAATVVINRRALRHNLQRLRELAPASKMVAVVKANAYGHGLLETARTLP
DADAFGVARLEEALRLRAGGITKPVLLLEGFFDARDLPTISAQHFHTAVH
NEEQLAALEEASLDEPVTVWMKLDTGMHRLGVRPEQAEAFYHRLTQCKNV
RQPVNIVSHFARADEPKCGATEKQLAIFNTFCEGKPGQRSIAASGGILLW
PQSHFDWVRPGIILYGVSPLEDRSTGADFGCQPVMSLTSSLIAVREHKAG
4.4Gene Sequence Similarity Search:
You will get several most similar results.
sequence format:
sequence format:
>gene fragment
ACAAAATCATCGTTGCTGACTGGAAGGTCGCCGACGCTGGTGAAACGCTCGCGCAACAG
GCGTTTGGCGCAGGCGCTAACTGGATGACCATCATCTGCGCCGCGCCGCTCGCGACGGT
AGAAAAAGGCCACGCAATGGCACAACGCTGCGGGGGTGAAATTCAGATAGAGCTGTTCG
GTAACTGGACGCTGGACGACGCCCGCGACTGG\nCATCGTATTGGCGTGCGGCAGGCCA
Z-score: score via FASTA algorithm.ACAAAATCATCGTTGCTGACTGGAAGGTCGCCGACGCTGGTGAAACGCTCGCGCAACAG
GCGTTTGGCGCAGGCGCTAACTGGATGACCATCATCTGCGCCGCGCCGCTCGCGACGGT
AGAAAAAGGCCACGCAATGGCACAACGCTGCGGGGGTGAAATTCAGATAGAGCTGTTCG
GTAACTGGACGCTGGACGACGCCCGCGACTGG\nCATCGTATTGGCGTGCGGCAGGCCA
5. Network Analysis
Node degree ranking(Top 10):
ten most complicated nodes, this may mean they are most important!
5.1 Name: the name of nodes
5.2 Degree: the degree of a node
5.3 Degree centrality: the degree centrality for nodes.
5.4 Density of graph: the density of graph
5.1 Name: the name of nodes
5.2 Degree: the degree of a node
5.3 Degree centrality: the degree centrality for nodes.
5.4 Density of graph: the density of graph
6. Data Processing Method
Some examples are used to explain data processing method
6.1 Keep main reaction(s), example 1:(R)-pantothenate + ATP --> D-4'-phosphopantothenate + ADP + H+
Obviously, ATP, ADP and H+ are current compounds (that is, they are not main compounds in this reaction), so we only keep (R)-pantothenate --> D-4'-phosphopantothenate.
Why? a reaction( A + B --> C + D ), according to graph theory, compounds(A, B, C, D) are nodes, reactions are edges in the graph. for example(a special reaction)
2 5-amino-levulinate --> porphobilinogen + H+ + 2 H2O
We can get three chemical equations from this reaction:
(1) 5-amino-levulinate --> porphobilinogen
(2) 5-amino-levulinate --> H+
(3) 5-amino-levulinate --> H2O
If we keep all of them, equation (2) and equation (3) make no sense. So we only keep main reaction(s) when processing data.
6.2 Struture considered, example 2:
L-aspartate + 2-oxoglutarate <--> oxaloacetate + L-glutamate
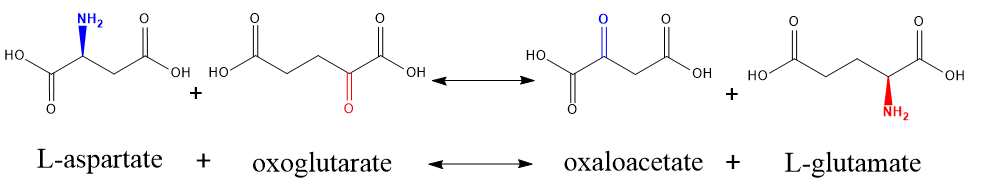
As is known to all, the reaction is a transaminatin. In transamination, the NH2 group on one molecule is exchanged with the =O group on the other molecule. The amino acid becomes a keto acid, and the keto acid becomes an amino acid. If we regard this reaction as an irreversible reaction, according to the picture above, struture considered, L-aspartate is similar to oxaloacetate (and oxoglutarate is similar to L-glutamate), so in this reaction L-aspartate is converted to oxaloacetate (and oxoglutarate is converted to L-glutamate) and we keep them as main reactions.
References:
1. KEGG reaction search: http://www.kegg.jp/dbget-bin/www_bfind_sub?&dbkey=reaction2. Albert, R. and Barabasi, A.L. (2002) Statistical mechanics of complex networks. Reviews of Modern Physics, 74, 47-97.
3. Albert, R., Barabasi, A.L. and Jeong, H. (2000) Power-law distribution of the World Wide Web. Science, 287, 2115.
4. Hongwu Ma and An-Ping Zeng. (2003) Reconstruction of metabolic networks from genome data and analysis of their global structure for various organisms. Bioinformatics, 19, 270-277.
5. Hongwu Ma and An-Ping Zeng. (2003) The connectivity structure, giant strong component and centrality of metabolic networks. Bioinformatics, 19, 1423-1430.
6. Neidhardt, F.C., Ingraham, J.L. and Schaechter, M. (1990) Physiology of the Bacterial Cell: a Molecular Approach. Sinauer Associates.