Background
Fermentation products, together with protogenetic food components, determine the sense, nutrition, and safety of fermented foods. Traditional “wet” experiment metabolite identification technique is time-consuming and cumbersome to operate and cannot meet the increasing need for identification of the extensive metabolites produced during food fermentation. Hence, we propose a data-driven strategy to explore microbial biosynthetic potential from genome scale and predict novel metabolites in food fermentation process. A food fermentation explorer was built based on state-of-the-art algorithms and data on 2,192,862 microbial sequenced- encoded enzymes. Using FFExplorer, we explained the mechanism behind the disappearance of spicy taste during pepper fermentation and the potential risks of fermented bean products. FFExplorer will provide a valuable reference for inferring bioactive “dark matter” in fermented foods and exploring the fermentation potential of microorganisms.
You can browse the strains used in FFExplorer from here and the biotransformation patterns here.
FFExplorer Implementation
FFExplorer was developed on the basis of the Linux, Apache, MySQL, Python (LAMP) strategy; the server runs under apache2 on a Linux machine running Ubuntu Server. The algorithm and backend program were written in Python by using the Django framework in combination with MySQL to manage the data. Hypertext Markup Language (HTML), Cascading Style Sheets (CSS), and JavaScript were used to implement FFExplorer’s front-end data presentation and interactions.
Model Parameters
SMILES The molecualr structure of fermentation substrate molecules. It can be entered into the prediction model in SMILES format. You can also enter the molecule into the prediction model via the JSME molecule editor.
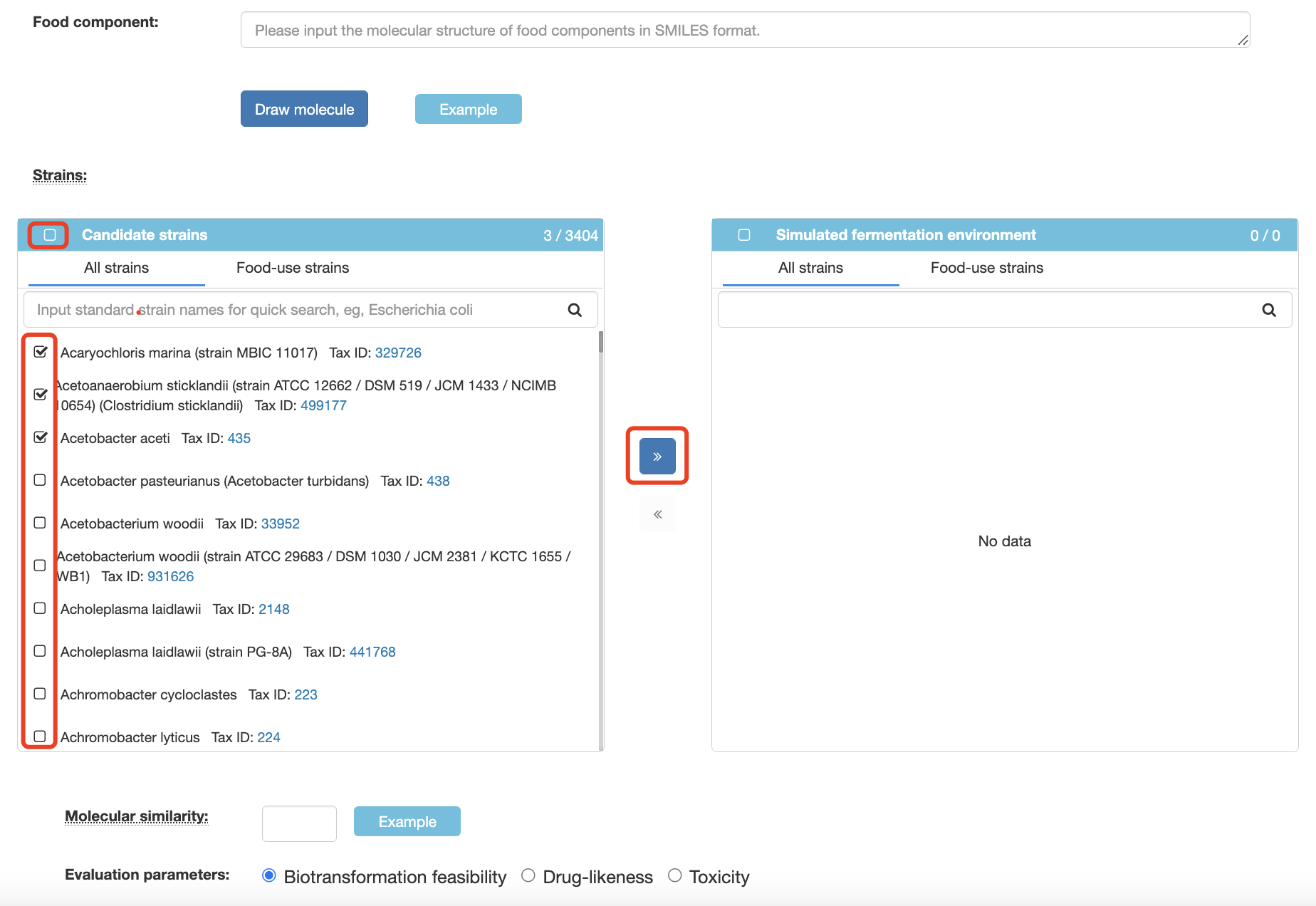
Strains You need to select one or more strains from the list of candidate strains by clicking the boxes in front of their names. Then a blue button will show up (looks like ">>"), you can add selected strains to the simulated fermentation environment by clicking the button.
You can also add all strains to the simulated fermentation environment at the same time to get more comprehensive predicted results by clicking the box in front of "Candidate strains".Molecular similarity The molecular similarity reflects the similarity between the inferred products and the pre-fermentation reactants, and it is calculated based on the maximum common substructure algorithm.
Evaluation parameters In order to evaluate the biological activity and synthesis feasibility of the inferred fermentation products, we introduced three indicators, including LD50, drug-likeness, synthetic assessment (SA) scores. LD50 and drug-likeness were predicted based on the ADMETLAB, which was used to evaluate the biological activity of the inferred fermentation product. The SA score was used to evaluate the difficulty of the fermentation product synthesis, which is obtained based on fragment contribution and complexity penalty. A lower SA score indicates that the fermented product is more easy to synthesise.
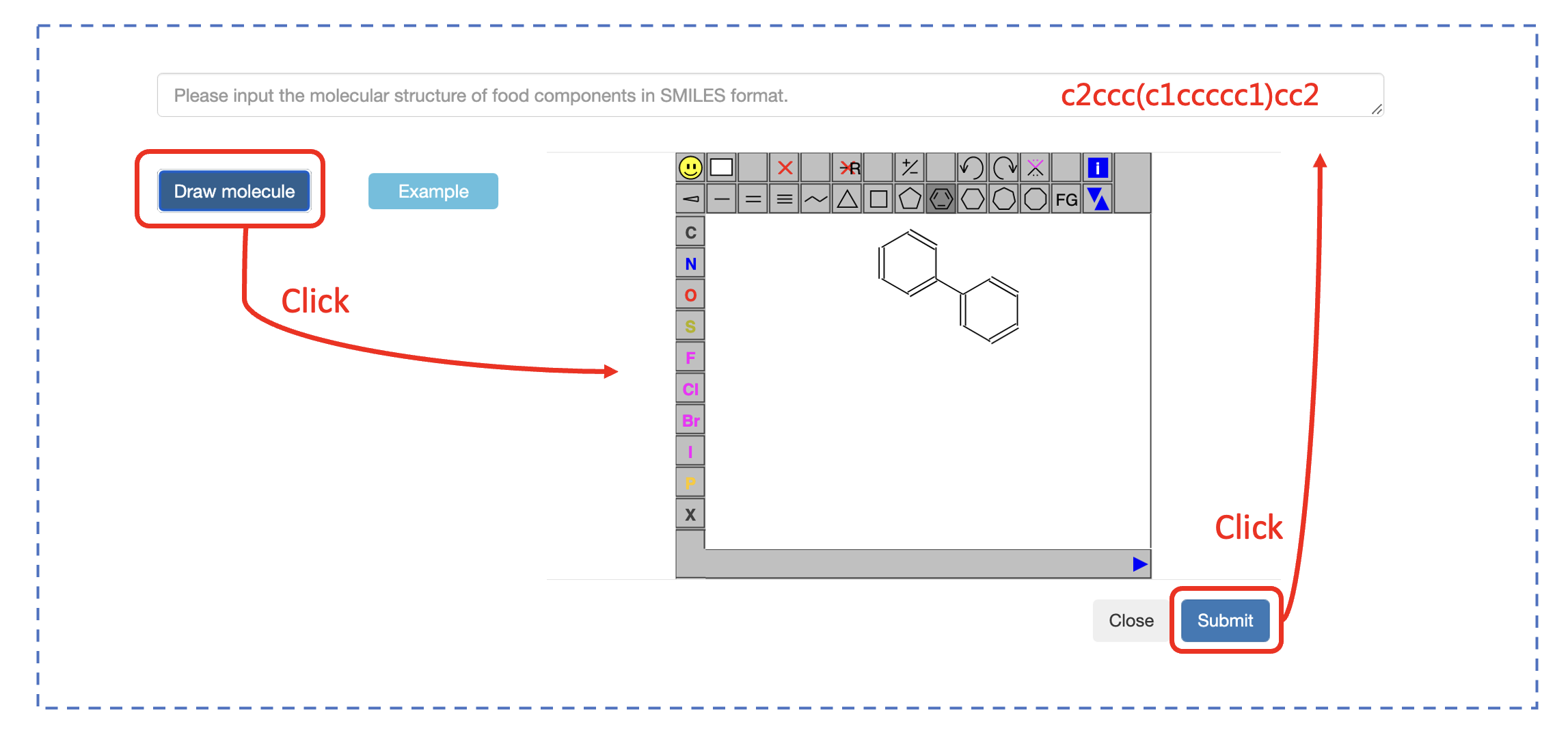
How to use FFExplorer
The first step is to input an active food ingredient to FFExplorer as the fermentation substrate. The molecular structure can be entered in SMILES format or via the JSME molecule editor, which have been integrated with FFExplorer. The second step is to choose fermentation microorganisms; the FFExplorer provides >3400 kinds of candidate microorganisms and >200 kinds of food-use microorganisms for the user to choose. For example, users can enter “Aspergillus” in the query bar in the list of candidate strains. All microorganisms contained “Aspergillus” in their scientific names will be displayed. Then users can select the microorganisms of interest by clicking the boxes in front of their names, then click the arrow button (looks like ">>") to add these microorganisms to the simulated fermentation environment. The third step is to specify the parameters, including the similarity, SA, LD50, and drug-likeness scores. Finally, clicking “Submit” to start the prediction. FFExplorer will automatically screen out the biotransformation patterns of the microorganisms in the simulated fermentation environment and use these biotransformation patterns to infer the possible structural modification of fermentation substrates. After a few minutes, FFExplorer will yield a tree diagram of the fermentation substrate and all predicted metabolites.
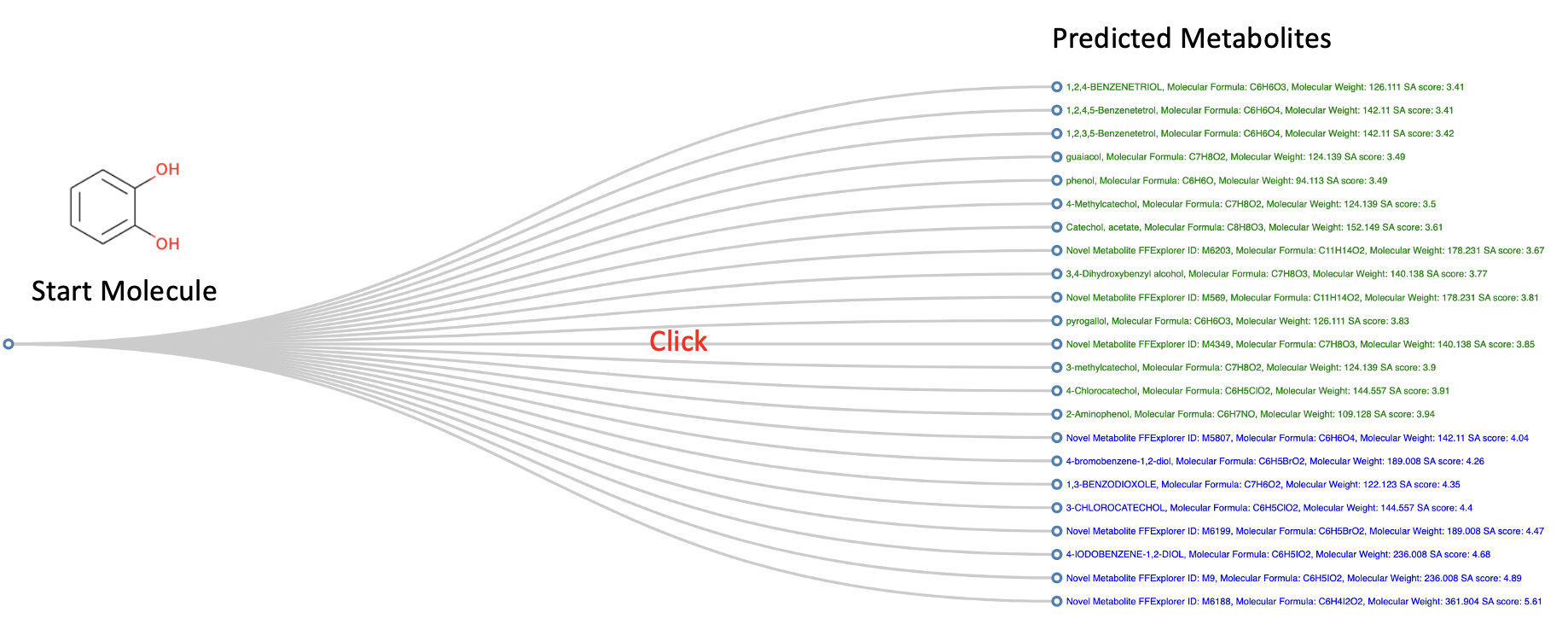
Upon clicking on the connection between the substrate and the metabolite, the FFExplorer will present the predicted metabolic pathways and referential reactions with similar biotransformation patterns.

Frequently asked questions
FFExplorer is offered to the public as a freely available resource. Use and re-distribution of the data, in whole or in part, not requires permission of the authors.
Please send an error report to zhangdachuan@picb.ac.cn and we will verify it as soon as possible. It would be very helpful if you could provide the correct information.
We recommend using Microsoft Edge. You can also use the latest version of other browsers.
If you have any other questions, please feel free to contact us by email zhangdachuan@picb.ac.cn.