Frequently Asked Questions
ToxinDB Implementation
The ToxinDB was implemented using PostgreSQL database engine, while all the scripts were written in Python. JSME were used for input molecular structure.
Data Availability
ToxinDB is offered to the public as a freely available resource. Use and re-distribution of the data not requires permission of the authors.
Multiple download interfaces have been developed to facilitate users to obtain data.
Submit Query
To satisfy the requirement of scientists being able to search for a specific molecular fragment or retrieve a class of toxins with structural similarities, the ToxinDB offers multiple retrieval methods, including full structure, fragment, similarity, maximum common substructure (MCS), and text retrieval options.
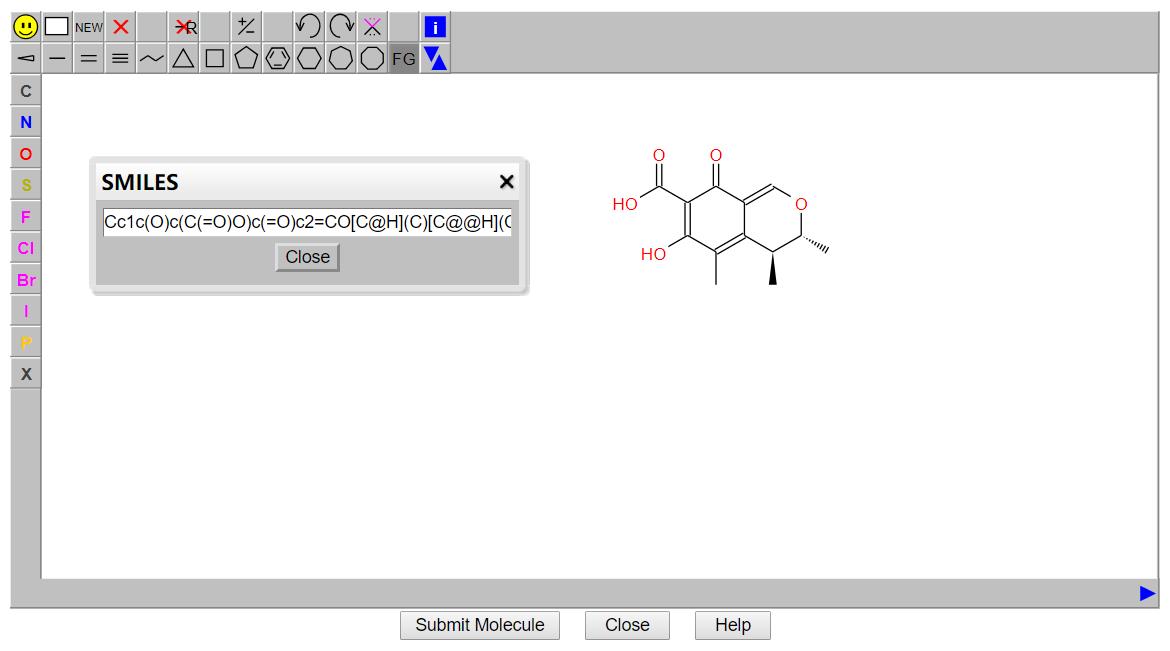
The JSME structure editor was included in the search options to enable searching through the drawing and editing of molecules.
Biotransformation Prediction
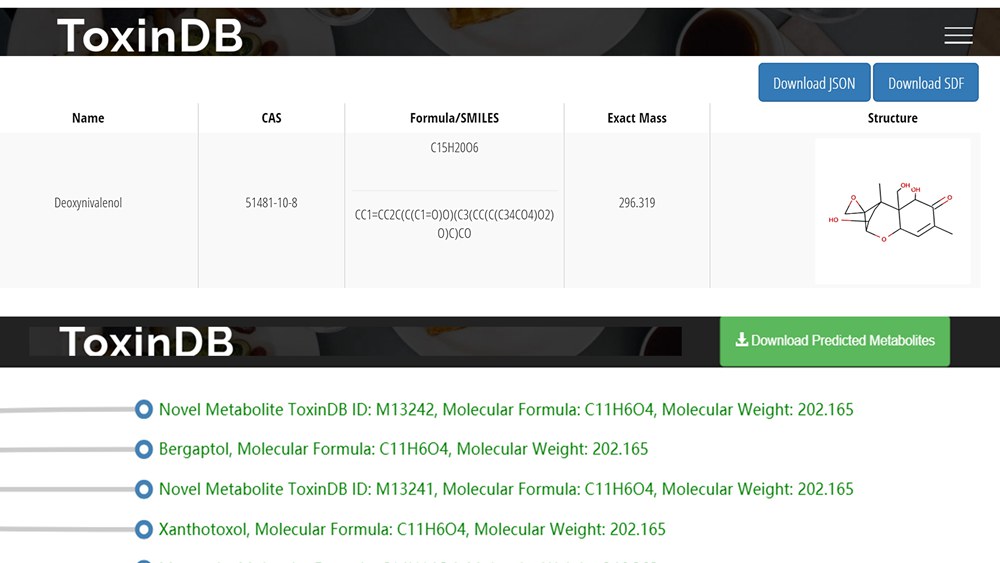
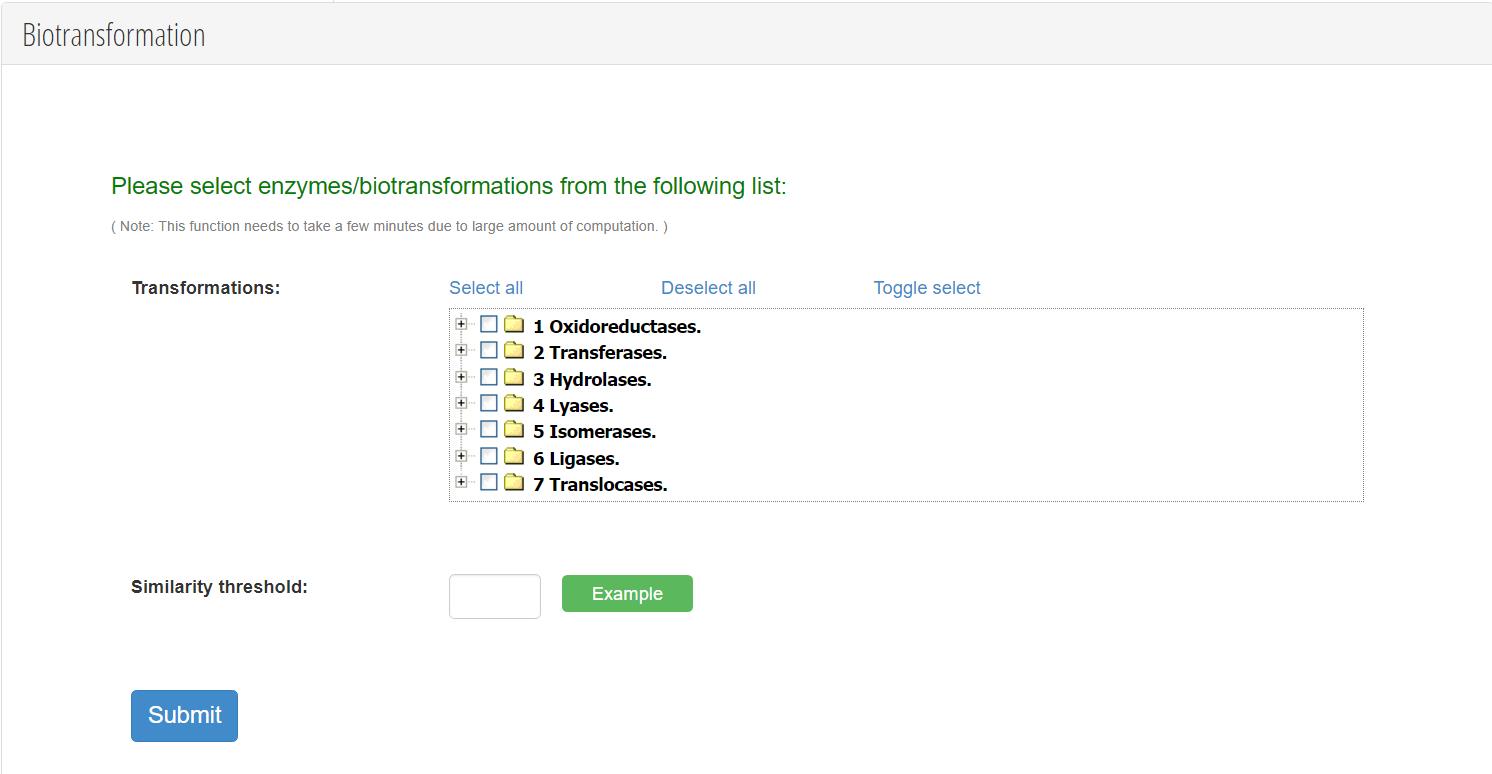
Using the approach described in manuscript, we extracted more than 8,000 unique biotransformation reaction rules. Each reaction rule represents a special enzyme's biotransformation ability. A prediction algorithm based on reaction rules was made into a user-friendly tool integrated within ToxinDB. One or more types of reaction rules can be selected when predicting toxin metabolites, for example oxidation (such as dehydrogenation of C-C bonds, epoxidation, hydroxylation), reduction (such as reduction of aldehydes, organic acids, ketones), hydrolysis (hydrolysis of amines, esters, lactones), or isomerisation (rearrangements, formation of C-C bonds or hetero-atom bonds). When searching for a toxin-detoxifying enzyme, scientists may select hydrolysis reaction rules, which are less demanding in the degradation environment and more suitable for practical use. In predicting toxin biomarkers, scientists can choose the phase I enzymes (mainly oxidoreductase) or the phase II enzymes (mainly transferase) that are expressed in the human body. Users can independently set the similarity threshold to filter the predicted results. The higher the set similarity threshold, the higher the feasibility of the predicted biotransformation. The prediction results were displayed in the form of a tree diagram. To facilitate scientists in distinguishing which predicted metabolites are reported compounds and which are novel, ToxinDB will automatic search predicted metabolites in PubChem database. If the predicted metabolite exists in the PubChem database, its compound name will be displayed. Otherwise, the system will mark it as ‘novel metabolite’.
Other Queries
Full structure retrieval means that a structural segment entered into the search by the user is fully consistent with the retrieved hit segment. To accelerate this process, we had previously normalized the molecular structure of all molecules in the ToxinDB. The molecular structure to be retrieved was normalized in real time and then matched to all normalized molecular structures in the ToxinDB, and molecules matching the exact structure were retrieved. Fragment retrieval (substructure retrieval) means that the submitted structure is a fragment and is therefore a connected subgraph of the hit structure. The similarity retrieval function employs a Tanimoto coefficient-based similarity algorithm. The retrieved molecular structure was iteratively compared to the molecules stored in the database, and the search eventually returned the 20 most similar toxins. The MCS retrieval function was based on the fMCS algorithm . This retrieval method differs from similarity retrieval in that it focuses more on the complete matching of partial structural fragments rather than the similarity of the overall structure. A text retrieval method is also available for searches based on the name, CAS number, InChI, InChI key, IUPAC name, or synonyms. The ToxinDB includes more than 100,000 synonyms of toxins to ensure precise matching to the query molecule.
ToxinDB is offered to the public as a freely available resource. Use and re-distribution of the data not requires permission of the authors. We encourage that users cite the ToxinDB paper in any resulting publications.
Please send an error report to zhangdachuan@picb.ac.cn and we will verify and correct it as soon as possible. Please list the references for the correct information if you can. That would be very helpful.
We recommend using Google Chrome. You can also use the latest version of other browsers.
If you have any other questions, please feel free to contact me with email qnhu@sibs.ac.cn.